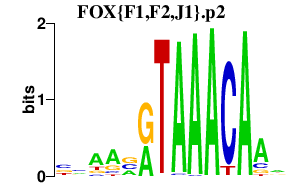
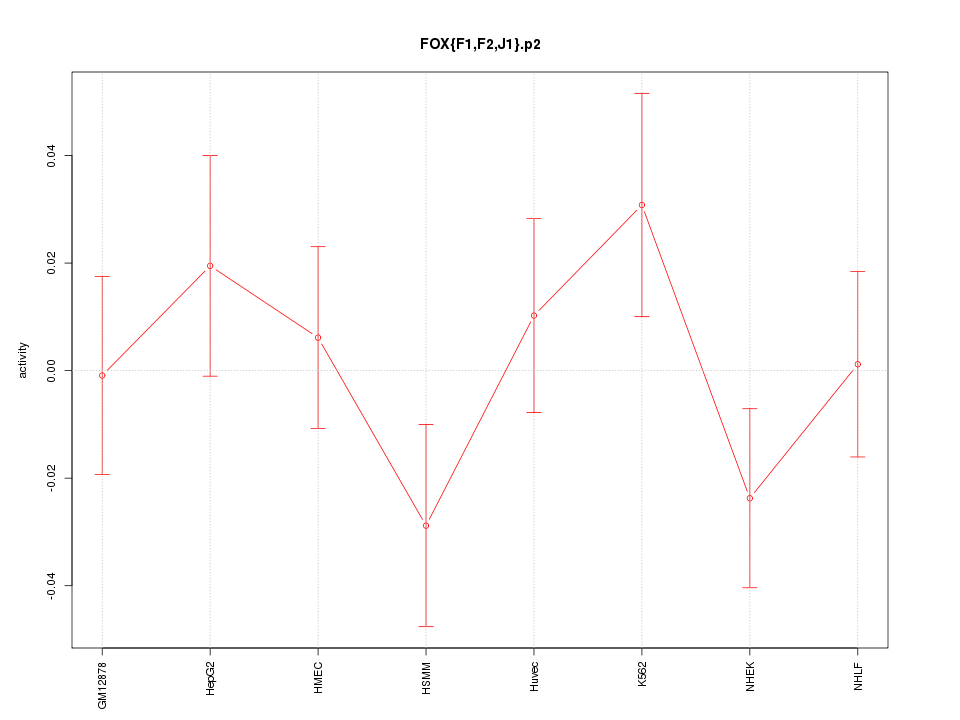
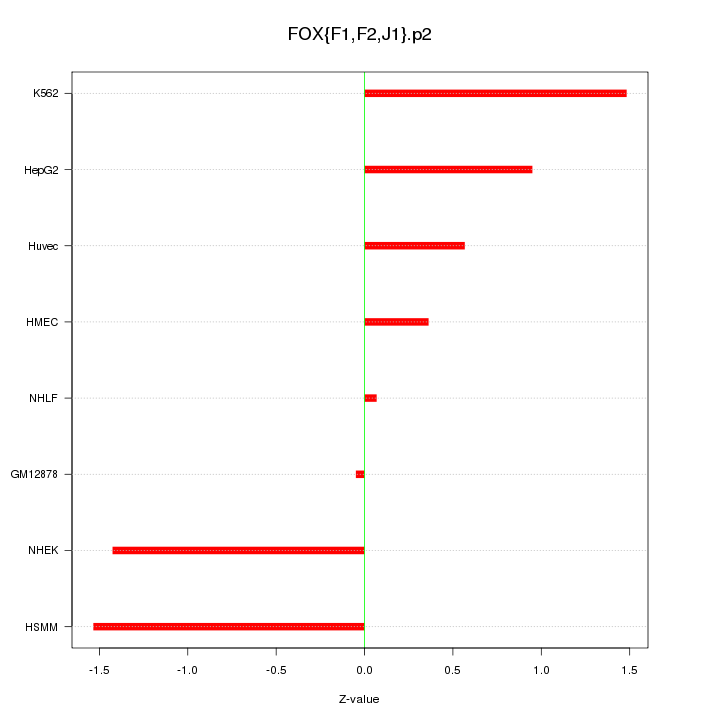
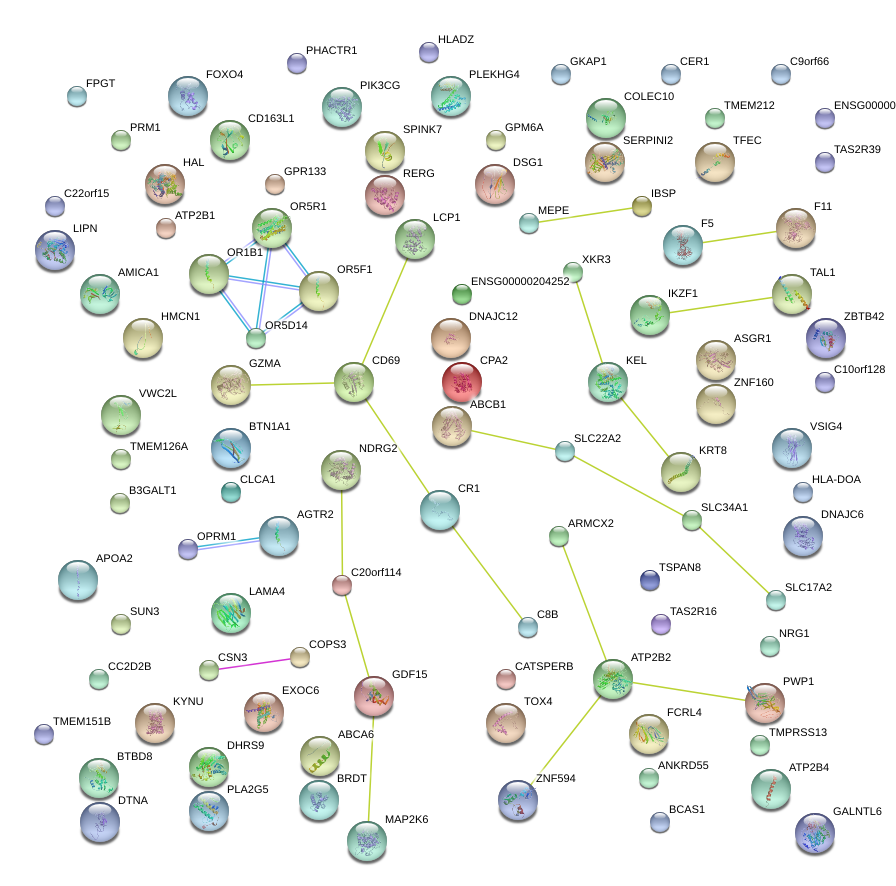
|
chr1_-_47468029
|
1.446
|
NM_003189
|
TAL1
|
T-cell acute lymphocytic leukemia 1
|
|
chrX_-_65176564
|
1.356
|
NM_001100431
NM_001184830
NM_001184831
NM_007268
|
VSIG4
|
V-set and immunoglobulin domain containing 4
|
|
chr22_-_15682555
|
1.117
|
NM_175878
|
XKR3
|
XK, Kell blood group complex subunit-related family, member 3
|
|
chr9_-_14712669
|
1.023
|
NM_005454
|
CER1
|
cerberus 1, cysteine knot superfamily, homolog (Xenopus laevis)
|
|
chr16_-_85099941
|
0.898
|
|
LOC400550
|
hypothetical LOC400550
|
|
chr13_-_45654327
|
0.870
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr13_-_45654296
|
0.853
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr13_-_45654292
|
0.838
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr19_+_18357966
|
0.835
|
NM_004864
|
GDF15
|
growth differentiation factor 15
|
|
chr11_-_104274606
|
0.794
|
NM_001191016
|
CASP12
|
caspase 12 (gene/pseudogene)
|
|
chr4_-_176971175
|
0.785
|
NM_201591
|
GPM6A
|
glycoprotein M6A
|
|
chr7_+_106292975
|
0.764
|
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr10_-_69267774
|
0.677
|
NM_021800
NM_201262
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12
|
|
chr9_-_205892
|
0.673
|
NM_152569
|
C9orf66
|
chromosome 9 open reading frame 66
|
|
chr6_-_112682573
|
0.663
|
|
LAMA4
|
laminin, alpha 4
|
|
chr7_+_129693938
|
0.640
|
NM_001869
|
CPA2
|
carboxypeptidase A2 (pancreatic)
|
|
chr1_-_159459999
|
0.621
|
NM_001643
|
APOA2
|
apolipoprotein A-II
|
|
chr7_+_106293112
|
0.615
|
NM_002649
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr1_+_183970093
|
0.606
|
NM_031935
|
HMCN1
|
hemicentin 1
|
|
chr2_+_168383427
|
0.600
|
NM_020981
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1
|
|
chr9_-_124431634
|
0.588
|
NM_001004450
|
OR1B1
|
olfactory receptor, family 1, subfamily B, member 1
|
|
chr14_-_20562962
|
0.541
|
NM_201538
NM_201539
NM_201540
NM_201541
|
NDRG2
|
NDRG family member 2
|
|
chr9_-_85622366
|
0.538
|
NM_001135953
NM_025211
|
GKAP1
|
G kinase anchoring protein 1
|
|
chr10_-_50066386
|
0.531
|
NM_001010863
|
C10orf128
|
chromosome 10 open reading frame 128
|
|
chr12_-_69837831
|
0.527
|
|
TSPAN8
|
tetraspanin 8
|
|
chr17_+_21946563
|
0.525
|
NM_001190452
|
MTRNR2L1
|
MT-RNR2-like 1
|
|
chr18_+_27152049
|
0.510
|
NM_001942
|
DSG1
|
desmoglein 1
|
|
chr14_-_91268136
|
0.510
|
NM_024764
|
CATSPERB
|
cation channel, sperm-associated, beta
|
|
chr1_+_65503011
|
0.509
|
NM_014787
|
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr7_-_122422989
|
0.502
|
NM_016945
|
TAS2R16
|
taste receptor, type 2, member 16
|
|
chr7_+_142590633
|
0.499
|
NM_176881
|
TAS2R39
|
taste receptor, type 2, member 39
|
|
chr3_+_46628928
|
0.495
|
NM_001195442
|
LOC100132146
|
hypothetical LOC100132146
|
|
chr4_+_187424111
|
0.494
|
NM_000128
|
F11
|
coagulation factor XI
|
|
chr2_+_169637255
|
0.494
|
NM_001142270
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr17_-_64649608
|
0.493
|
NM_080284
|
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6
|
|
chr11_-_55942283
|
0.490
|
NM_001004744
|
OR5R1
|
olfactory receptor, family 5, subfamily R, member 1
|
|
chr1_-_57204208
|
0.489
|
NM_000066
|
C8B
|
complement component 8, beta polypeptide
|
|
chr16_-_11282595
|
0.485
|
NM_002761
|
PRM1
|
protamine 1
|
|
chr5_+_147672181
|
0.476
|
NM_032566
|
SPINK7
|
serine peptidase inhibitor, Kazal type 7 (putative)
|
|
chr6_-_112682401
|
0.476
|
|
LAMA4
|
laminin, alpha 4
|
|
chr4_+_88973144
|
0.474
|
NM_001184695
NM_001184696
NM_001184697
NM_020203
|
MEPE
|
matrix extracellular phosphoglycoprotein
|
|
chr4_+_187424336
|
0.474
|
|
F11
|
coagulation factor XI
|
|
chr6_+_21774624
|
0.474
|
|
FLJ22536
|
hypothetical locus LOC401237
|
|
chr1_+_74473672
|
0.459
|
NM_015978
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chr11_-_117305277
|
0.452
|
NM_001077263
|
TMPRSS13
|
transmembrane protease, serine 13
|
|
chr12_-_15265490
|
0.452
|
NM_001190726
NM_032918
|
RERG
|
RAS-like, estrogen-regulated, growth inhibitor
|
|
chr7_-_48035169
|
0.434
|
NM_001030019
NM_152782
|
SUN3
|
Sad1 and UNC84 domain containing 3
|
|
chr1_+_20269239
|
0.433
|
NM_000929
|
PLA2G5
|
phospholipase A2, group V
|
|
chr14_+_104338423
|
0.431
|
|
ZBTB42
|
zinc finger and BTB domain containing 42
|
|
chr4_+_172971149
|
0.431
|
NM_001034845
|
GALNTL6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6
|
|
chr1_+_86707113
|
0.430
|
NM_001285
|
CLCA1
|
chloride channel accessory 1
|
|
chr6_-_160599779
|
0.426
|
NM_003058
|
SLC22A2
|
solute carrier family 22 (organic cation transporter), member 2
|
|
chr6_-_33085290
|
0.425
|
|
HLA-DOA
|
major histocompatibility complex, class II, DO alpha
|
|
chr6_+_12825818
|
0.417
|
NM_030948
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr3_-_10522267
|
0.416
|
NM_001001331
NM_001683
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr5_+_54434230
|
0.410
|
NM_006144
|
GZMA
|
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3)
|
|
chr2_+_143351695
|
0.410
|
|
KYNU
|
kynureninase
|
|
chr2_+_214984705
|
0.408
|
NM_001080500
|
VWC2L
|
von Willebrand factor C domain containing protein 2-like
|
|
chr22_-_16127520
|
0.407
|
|
CECR3
|
cat eye syndrome chromosome region, candidate 3
|
|
chr12_-_51629874
|
0.401
|
|
KRT8
|
keratin 8
|
|
chr16_+_65868913
|
0.401
|
NM_015432
|
PLEKHG4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4
|
|
chrX_+_70232362
|
0.399
|
NM_001170931
NM_005938
|
FOXO4
|
forkhead box O4
|
|
chr7_-_115458010
|
0.397
|
NM_001018058
NM_012252
|
TFEC
|
transcription factor EC
|
|
chr6_+_154373323
|
0.392
|
NM_001145279
NM_001145280
NM_001145281
|
OPRM1
|
opioid receptor, mu 1
|
|
chr12_-_7487989
|
0.382
|
NM_174941
|
CD163L1
|
CD163 molecule-like 1
|
|
chr6_+_44352443
|
0.376
|
|
TMEM151B
|
transmembrane protein 151B
|
|
chr10_+_97749837
|
0.376
|
NM_001001732
NM_001159747
|
CC2D2B
|
coiled-coil and C2 domain containing 2B
|
|
chr12_+_130004404
|
0.368
|
NM_198827
|
GPR133
|
G protein-coupled receptor 133
|
|
chr3_+_173043832
|
0.367
|
NM_001164436
|
TMEM212
|
transmembrane protein 212
|
|
chr1_+_92318449
|
0.366
|
NM_183242
|
BTBD8
|
BTB (POZ) domain containing 8
|
|
chr3_-_10522340
|
0.364
|
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr4_+_71142893
|
0.361
|
NM_005212
|
CSN3
|
casein kappa
|
|
chr11_-_117589283
|
0.360
|
NM_153206
|
AMICA1
|
adhesion molecule, interacts with CXADR antigen 1
|
|
chr10_+_90511142
|
0.354
|
NM_001102469
|
LIPN
|
lipase, family member N
|
|
chr6_+_26609427
|
0.351
|
NM_001732
|
BTN1A1
|
butyrophilin, subfamily 1, member A1
|
|
chr6_+_62341966
|
0.350
|
NM_001190706
|
MTRNR2L9
|
MT-RNR2-like 9
|
|
chr7_+_50318801
|
0.350
|
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr7_+_106292932
|
0.349
|
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr22_+_22435207
|
0.348
|
NM_182520
|
C22orf15
|
chromosome 22 open reading frame 15
|
|
chr5_-_55564942
|
0.344
|
NM_024669
|
ANKRD55
|
ankyrin repeat domain 55
|
|
chr7_+_142053693
|
0.344
|
NM_001190487
|
MTRNR2L6
|
MT-RNR2-like 6
|
|
chr1_+_92190296
|
0.343
|
NM_001726
|
BRDT
|
bromodomain, testis-specific
|
|
chr12_-_94914133
|
0.342
|
NM_002108
|
HAL
|
histidine ammonia-lyase
|
|
chr8_+_120148604
|
0.339
|
NM_006438
|
COLEC10
|
collectin sub-family member 10 (C-type lectin)
|
|
chr2_+_88966170
|
0.335
|
|
|
|
|
chr6_-_26038817
|
0.335
|
NM_005835
|
SLC17A2
|
solute carrier family 17 (sodium phosphate), member 2
|
|
chr20_+_31334601
|
0.334
|
NM_033197
|
C20orf114
|
chromosome 20 open reading frame 114
|
|
chr11_+_55319607
|
0.333
|
NM_001004735
|
OR5D14
|
olfactory receptor, family 5, subfamily D, member 14
|
|
chr18_+_30327243
|
0.333
|
NM_001198945
NM_001392
NM_032975
NM_032979
|
DTNA
|
dystrobrevin, alpha
|
|
chr1_+_205736095
|
0.332
|
NM_000573
NM_000651
|
CR1
|
complement component (3b/4b) receptor 1 (Knops blood group)
|
|
chr1_-_155834357
|
0.331
|
NM_031282
|
FCRL4
|
Fc receptor-like 4
|
|
chr12_-_9804682
|
0.330
|
NM_001781
|
CD69
|
CD69 molecule
|
|
chr7_-_87180499
|
0.325
|
NM_000927
|
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1
|
|
chr4_+_88939725
|
0.324
|
NM_004967
|
IBSP
|
integrin-binding sialoprotein
|
|
chr17_-_7023378
|
0.323
|
NM_001197216
NM_001671
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr5_-_42847716
|
0.321
|
NM_001085486
NM_001093726
NM_005410
|
SEPP1
|
selenoprotein P, plasma, 1
|
|
chrX_+_115215985
|
0.321
|
NM_000686
|
AGTR2
|
angiotensin II receptor, type 2
|
|
chr11_-_55518675
|
0.318
|
NM_003697
|
OR5F1
|
olfactory receptor, family 5, subfamily F, member 1
|
|
chr1_-_167822248
|
0.316
|
|
F5
|
coagulation factor V (proaccelerin, labile factor)
|
|
chr3_-_168674502
|
0.316
|
NM_006217
|
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2
|
|
chr10_+_94584449
|
0.315
|
NM_001013848
|
EXOC6
|
exocyst complex component 6
|
|
chr20_-_43996980
|
0.312
|
|
FLJ40606
|
hypothetical protein LOC643549
|
|
chr17_+_64922420
|
0.310
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr20_-_52120421
|
0.308
|
|
BCAS1
|
breast carcinoma amplified sequence 1
|
|
chr7_-_142369519
|
0.304
|
|
KEL
|
Kell blood group, metallo-endopeptidase
|
|
chr11_-_5178505
|
0.303
|
NM_001004760
|
OR51V1
|
olfactory receptor, family 51, subfamily V, member 1
|
|
chr20_+_16676997
|
0.302
|
NM_020157
|
OTOR
|
otoraplin
|
|
chr12_-_14929991
|
0.299
|
NM_000900
NM_001190839
|
MGP
|
matrix Gla protein
|
|
chr20_-_49612574
|
0.296
|
NM_001136021
|
NFATC2
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2
|
|
chr12_-_51332214
|
0.295
|
NM_000423
|
KRT2
|
keratin 2
|
|
chr2_+_143351536
|
0.294
|
NM_001032998
NM_003937
|
KYNU
|
kynureninase
|
|
chr1_-_244796169
|
0.294
|
NM_022366
|
TFB2M
|
transcription factor B2, mitochondrial
|
|
chr4_+_35962038
|
0.292
|
NM_001170700
|
DTHD1
|
death domain containing 1
|
|
chr1_+_150753601
|
0.292
|
NM_019060
|
CRCT1
|
cysteine-rich C-terminal 1
|
|
chr11_+_5925152
|
0.292
|
NM_001003443
|
OR56A3
|
olfactory receptor, family 56, subfamily A, member 3
|
|
chr4_+_91375204
|
0.292
|
NM_207491
|
FAM190A
|
family with sequence similarity 190, member A
|
|
chr20_+_51023159
|
0.287
|
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr7_-_107230867
|
0.282
|
NM_000111
|
SLC26A3
|
solute carrier family 26, member 3
|
|
chr19_-_38052481
|
0.281
|
NM_001126335
NM_014270
|
SLC7A9
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 9
|
|
chr5_-_180009205
|
0.280
|
|
FLT4
|
fms-related tyrosine kinase 4
|
|
chr2_+_161980865
|
0.280
|
NM_006593
|
TBR1
|
T-box, brain, 1
|
|
chr5_-_27074407
|
0.280
|
NM_016279
|
CDH9
|
cadherin 9, type 2 (T1-cadherin)
|
|
chr10_+_96512447
|
0.279
|
NM_000769
|
CYP2C19
|
cytochrome P450, family 2, subfamily C, polypeptide 19
|
|
chr7_-_140270749
|
0.278
|
NM_004333
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B1
|
|
chr18_+_30327341
|
0.277
|
|
DTNA
|
dystrobrevin, alpha
|
|
chr17_+_53587492
|
0.276
|
NM_012374
|
OR4D1
|
olfactory receptor, family 4, subfamily D, member 1
|
|
chr11_+_59027624
|
0.275
|
NM_001004706
|
OR4D11
|
olfactory receptor, family 4, subfamily D, member 11
|
|
chr18_+_59374372
|
0.272
|
NM_080474
|
SERPINB12
|
serpin peptidase inhibitor, clade B (ovalbumin), member 12
|
|
chr4_-_82612049
|
0.272
|
NM_152545
|
RASGEF1B
|
RasGEF domain family, member 1B
|
|
chr5_-_169657360
|
0.270
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr17_-_36276987
|
0.270
|
NM_000223
|
KRT12
|
keratin 12
|
|
chr2_-_162717159
|
0.270
|
NM_002054
|
GCG
|
glucagon
|
|
chr12_+_92595281
|
0.269
|
NM_003805
|
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain
|
|
chr6_-_2696152
|
0.269
|
NM_001012418
|
MYLK4
|
myosin light chain kinase family, member 4
|
|
chr5_-_20024050
|
0.268
|
NM_001167667
NM_004934
|
CDH18
|
cadherin 18, type 2
|
|
chr1_-_245763763
|
0.266
|
NM_198074
|
OR2C3
|
olfactory receptor, family 2, subfamily C, member 3
|
|
chr21_-_14840506
|
0.265
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr12_-_101835026
|
0.265
|
|
PAH
|
phenylalanine hydroxylase
|
|
chr8_-_16094591
|
0.264
|
NM_002445
NM_138715
NM_138716
|
MSR1
|
macrophage scavenger receptor 1
|
|
chr11_+_48194937
|
0.262
|
NM_001005470
|
OR4B1
|
olfactory receptor, family 4, subfamily B, member 1
|
|
chr5_-_110090264
|
0.261
|
NM_001039763
|
TMEM232
|
transmembrane protein 232
|
|
chr7_-_142369534
|
0.261
|
NM_000420
|
KEL
|
Kell blood group, metallo-endopeptidase
|
|
chr10_+_4995453
|
0.250
|
NM_001353
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr1_-_155788775
|
0.250
|
NM_001195388
NM_031281
|
FCRL5
|
Fc receptor-like 5
|
|
chr5_-_169657312
|
0.249
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr20_-_43609409
|
0.249
|
NM_001198986
NM_020398
|
SPINLW1-WFDC6
SPINLW1
|
SPINLW1-WFDC6 readthrough
serine peptidase inhibitor-like, with Kunitz and WAP domains 1 (eppin)
|
|
chr7_-_112513379
|
0.249
|
NM_001146266
NM_001146267
|
GPR85
|
G protein-coupled receptor 85
|
|
chr1_-_244796062
|
0.248
|
|
TFB2M
|
transcription factor B2, mitochondrial
|
|
chr11_-_102331643
|
0.248
|
NM_002427
|
MMP13
|
matrix metallopeptidase 13 (collagenase 3)
|
|
chr9_+_128764324
|
0.247
|
NM_001190728
|
RALGPS1
|
Ral GEF with PH domain and SH3 binding motif 1
|
|
chr2_-_21084077
|
0.247
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr15_+_56217686
|
0.246
|
NM_020980
|
AQP9
|
aquaporin 9
|
|
chr9_+_132990760
|
0.242
|
NM_005085
|
NUP214
|
nucleoporin 214kDa
|
|
chr11_+_60309396
|
0.242
|
NM_206893
|
MS4A10
|
membrane-spanning 4-domains, subfamily A, member 10
|
|
chr6_+_32229599
|
0.242
|
NM_005155
|
PPT2-EGFL8
PPT2
|
PPT2-EGFL8 read-through transcript
palmitoyl-protein thioesterase 2
|
|
chr7_-_25234582
|
0.242
|
NM_022150
|
NPVF
|
neuropeptide VF precursor
|
|
chr1_+_244796261
|
0.242
|
NM_001139459
NM_152609
|
CNST
|
consortin, connexin sorting protein
|
|
chr6_-_31740466
|
0.241
|
|
GPANK1
|
G patch domain and ankyrin repeats 1
|
|
chrX_-_15242589
|
0.241
|
NM_001012428
|
ASB11
|
ankyrin repeat and SOCS box containing 11
|
|
chr12_+_98565663
|
0.241
|
NM_153364
|
FAM71C
|
family with sequence similarity 71, member C
|
|
chr2_+_1396239
|
0.241
|
NM_000547
NM_175719
|
TPO
|
thyroid peroxidase
|
|
chr5_-_180009178
|
0.241
|
|
FLT4
|
fms-related tyrosine kinase 4
|
|
chr1_-_195302973
|
0.240
|
NM_001994
|
F13B
|
coagulation factor XIII, B polypeptide
|
|
chr19_+_15612706
|
0.240
|
NM_000896
|
CYP4F3
|
cytochrome P450, family 4, subfamily F, polypeptide 3
|
|
chr1_+_244796426
|
0.240
|
|
CNST
|
consortin, connexin sorting protein
|
|
chr4_+_74488910
|
0.239
|
|
ALB
|
albumin
|
|
chr15_-_38387266
|
0.238
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr6_+_136214495
|
0.236
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr16_+_11346780
|
0.234
|
NM_152308
|
C16orf75
|
chromosome 16 open reading frame 75
|
|
chr1_+_205344200
|
0.234
|
NM_000715
|
C4BPA
|
complement component 4 binding protein, alpha
|
|
chr12_-_61615124
|
0.233
|
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr6_+_32230172
|
0.233
|
|
PPT2
|
palmitoyl-protein thioesterase 2
|
|
chr7_-_126670804
|
0.233
|
NM_000845
|
GRM8
|
glutamate receptor, metabotropic 8
|
|
chr13_-_38462857
|
0.233
|
NM_001144033
NM_145286
|
STOML3
|
stomatin (EPB72)-like 3
|
|
chr1_+_244796441
|
0.232
|
|
CNST
|
consortin, connexin sorting protein
|
|
chr4_-_140443061
|
0.232
|
NM_001184987
NM_001184988
NM_001184989
NM_001184990
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa
|
|
chr20_+_30061905
|
0.229
|
NM_080625
|
C20orf160
|
chromosome 20 open reading frame 160
|
|
chr1_+_195503956
|
0.228
|
NM_001193640
NM_201253
|
CRB1
|
crumbs homolog 1 (Drosophila)
|
|
chr2_-_165133239
|
0.228
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr5_-_180009150
|
0.227
|
|
FLT4
|
fms-related tyrosine kinase 4
|
|
chr12_+_120055013
|
0.226
|
NM_002562
|
P2RX7
|
purinergic receptor P2X, ligand-gated ion channel, 7
|
|
chr12_-_11439752
|
0.225
|
NM_006248
|
PRB1
PRB2
|
proline-rich protein BstNI subfamily 1
proline-rich protein BstNI subfamily 2
|
|
chr5_-_169657395
|
0.224
|
NM_005565
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr18_+_53170718
|
0.224
|
NM_015879
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3
|
|
chr17_-_72151314
|
0.224
|
NM_018414
|
ST6GALNAC1
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1
|
|
chr15_-_38387357
|
0.224
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr1_+_245779018
|
0.223
|
NM_145278
|
C1orf150
|
chromosome 1 open reading frame 150
|
|
chr8_+_100025806
|
0.222
|
NM_001142462
NM_053001
|
OSR2
|
odd-skipped related 2 (Drosophila)
|
|
chr6_-_168222645
|
0.220
|
NM_024919
|
FRMD1
|
FERM domain containing 1
|
|
chr22_+_48951286
|
0.220
|
NM_001160300
NM_052839
|
PANX2
|
pannexin 2
|
|
chr4_-_155531898
|
0.218
|
NM_017639
|
DCHS2
|
dachsous 2 (Drosophila)
|
|
chr6_-_112682459
|
0.217
|
NM_001105206
NM_001105207
NM_001105208
NM_001105209
NM_002290
|
LAMA4
|
laminin, alpha 4
|
|
chr17_-_31104009
|
0.217
|
NM_139285
|
GAS2L2
|
growth arrest-specific 2 like 2
|
|
chr16_+_54782802
|
0.216
|
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O
|
|
chr4_-_100575313
|
0.215
|
NM_001166504
NM_000673
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide
|
|
chr7_+_113513828
|
0.213
|
|
FOXP2
|
forkhead box P2
|
|
chr20_-_22514092
|
0.212
|
NM_153675
|
FOXA2
|
forkhead box A2
|
|
chr7_-_93041971
|
0.212
|
NM_001164737
NM_001742
|
CALCR
|
calcitonin receptor
|
|
chr12_-_7709734
|
0.211
|
NM_001644
|
APOBEC1
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1
|
|
chr17_+_7180571
|
0.210
|
NM_014716
|
ACAP1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1
|